Modeling fiber confinement within cell monolayers in Cytosim¶
Author: Marco Pensalfini, UPC-BarcelonaTech, April 2022
Preamble¶
Monolayers are cohesive cellular sheets that appear in a number of physiological functions, whereby cell-cell junctions and cytoskeletal filaments allow integrating $-$ physicall and functionally $-$ individual cells into three-dimensional tissue membranes. While mainly interested in single-cell biophysics, we want to be able to constrain the modeled cytoskeletal fibers by accounting for idealized interactions with the neighboring cells and their cytoskeletal networks. To this end, we have developed a dedicated confining space in Cytosim, which represents a right regular prism and a set of custom-implemented fiber confinement options.
A right regular prism as confining space: the "polyreg" class¶
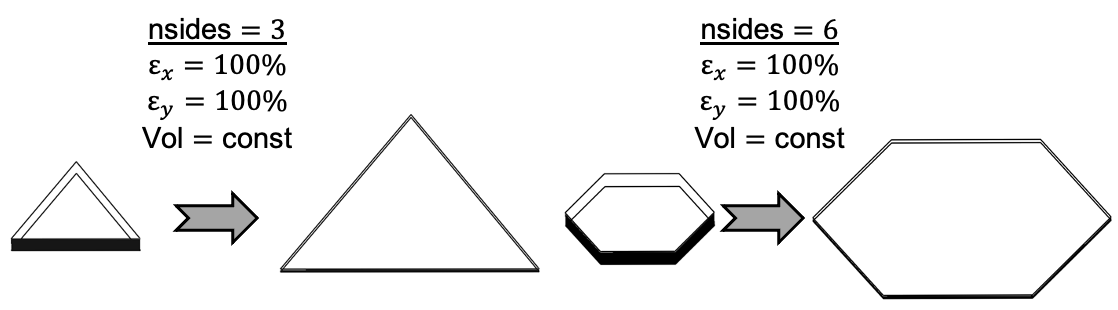
Owing to the typical membrane-like characteristics of cell monolayers, we developed a custom space class in Cytosim that corresponds to the extrusion of a regular polygon. This can be accessed through the keyword "shape = polyreg" and takes the following parameters:
- radius $\rightarrow$ the radius of the circle passing through the vertices of the polygonal base, i.e. the inscribing circle in the undeformed cell configuration;
- nsides $\rightarrow$ the number of sides of the polygonal base, which will all have the same length in the undeformed cell configuration;
- deforgrad $\rightarrow$ the 5 (4 in plane + 1 for thickness) components of the deformation gradient, describing the current deformation from the reference;
- height $\rightarrow$ the extrusion length, i.e. the cell height in the undeformed cell configuration.
Note that updating the value of "deforgrad" during a simulation will allow prescribing the cell deformation, e.g. to simulate mechanical stretching, as shown in the example images below.
Custom fiber confinement options for the "polyreg" class¶
In addition to the standard fiber confinement options, "polyreg" has a few additional capabilities that are useful to model a single cell as part of a monolayer by representing the following cases:
- fiber ends free to slide on the lateral cell walls, but prevented from leaving the wall where they are initially found
- fiber ends free to slide on the lateral cell walls
- fiber ends constrained to maintain their position on the lateral cell walls
In each of the above cases, all fiber points except for their extremities are confined inside the current volume of the cell (prism) in a manner that is equivalent to the standard "CONFINE_INSIDE".
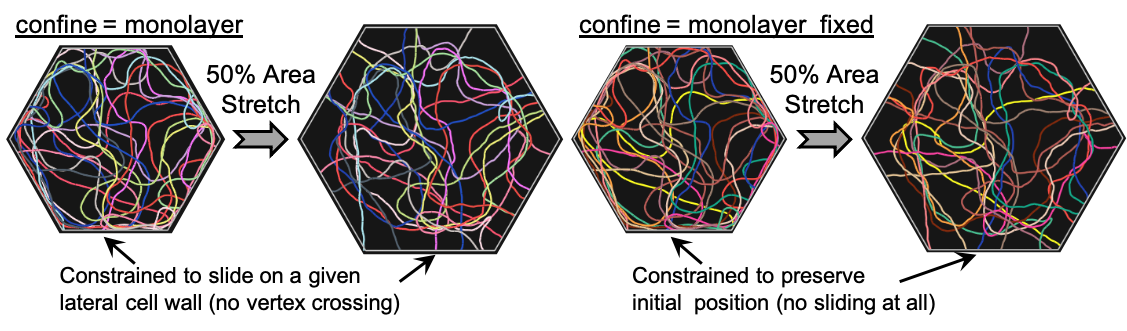
Case 1: confine = monolayer¶
As a first case, we consider a fiber whose ends are constrained to slide on the lateral cell walls. This idealizes the situation where cytoskeletal filaments within a cell are connected to those of neighboring cells through cell-cell junctions as part of a monolayer. Assuming that the membrane provides no resistance to sliding, we can further model the motion of filament ends on the lateral cell walls as frictionless and link the current location of each fiber end to its projection on a cell wall through a harmonic potential that vanishes at the projection point, as it is typical in Cytosim. Moreover, considering the connectivity between neighboring cells as fixed, we additionally limit the fiber end mobility to the cell side wall where they are found at the beginning of the simulation.
This situation can be modeled in Cytosim by providing the keyword "confine = monolayer" while defining the fiber properties, knowing that it takes up to 3 additional arguments (see example code a few lines below):
- the stiffness of the potential, which applies to all points comprising a fiber (ends slide on the membrane and all other points confined inside the cell);
- the name of the confining space (optional);
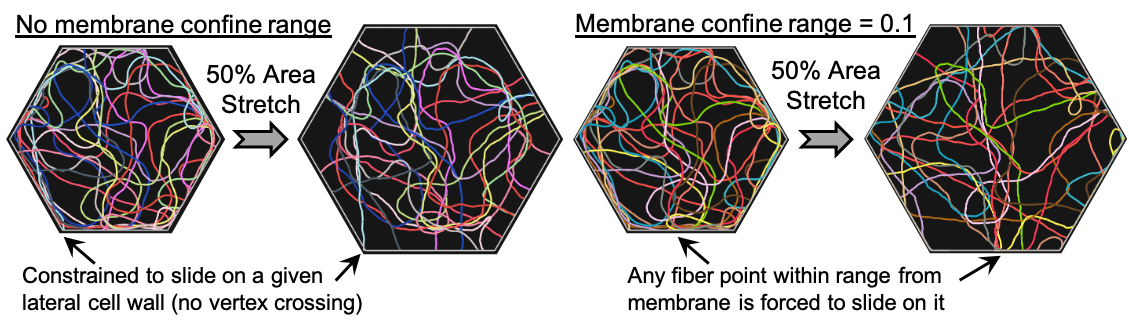
- the range of the membrane-binding action (optional), in which case all fiber points within this range will be constrained to slide frictionlessly on the membrane.
cytosim
set fiber filament
{
rigidity = 0.021
segmentation = 0.2
confine = monolayer, 1000.0, cell, 0.1
display = ( coloring = 1; )
}Below we showcase the effect of such a fiber confinement on the mobility of cytoskeletal filaments that are contained within a hexagonal cell undergoing 10% equibiaxial areal strain (see example file "filament_network_monolayer.cym").
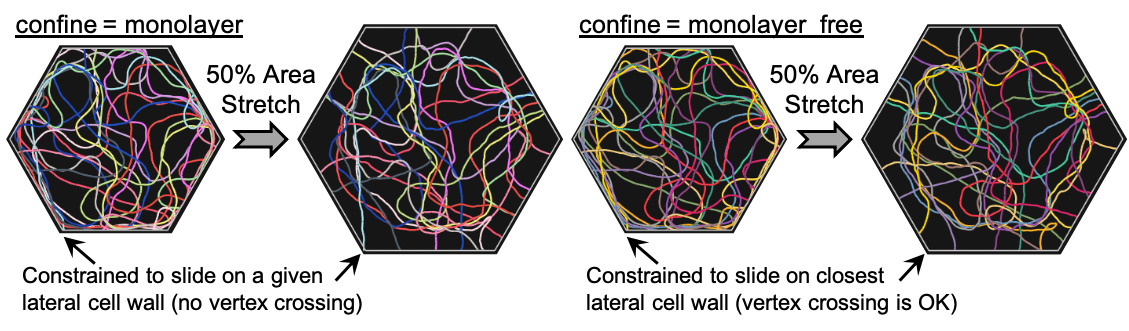
Case 2: confine = monolayer_free¶
As a limiting case, we consider letting the fiber ends slide freely on any lateral cell wall. The only difference from the previous case is that fiber points are now projected on the closest lateral cell wall, irrespective of their initial configuration.
This situation can also be modeled in Cytosim by replacing "confine = monolayer" with "confine = monolayer_free" and providing parameters that are analogous to those previously described.
Case 3: confine = monolayer_fixed¶
We conclude by considering a limiting case where the membrane-induced friction entirely prevents fiber ends from sliding. Each fiber end is now linked to its initial position using a harmonic potential of given stiffness; this situation can be modeled in Cytosim by using "confine = monolayer_fixed" and providing the usual parameters for monolayer confinement.