Controlling 2D fiber nematic ordering in Cytosim¶
Author: Marco Pensalfini, UPC-BarcelonaTech, April 2022
Objective¶
This tutorial discusses a custom-implemented 2D organizer class that allows controlling the nematic ordering of a fiber ensemble (e.g. actin) in Cytosim.
We will cover the theory and algorithms that underlie the implementation and provide some applicative examples. The code is available on GitLab as part of Cytosim.
Preamble¶
Nematic structures are ubiquitous cytoskeletal architectures that emerge from the organization of aligned actin filaments with mixed polarity in association with various binding proteins, e.g. myosin-II motors and crosslinks.
Nematics can be characterized using the nematic tensor, which in 2D reads $q_{ij} = S(n_in_j - \delta_{ij}/2)$ and features two parameters that are linked to actin orientation:
- $\mathbf{n}$ is the nematic director, a 2D unit vector that represents the main axis of nematic order;
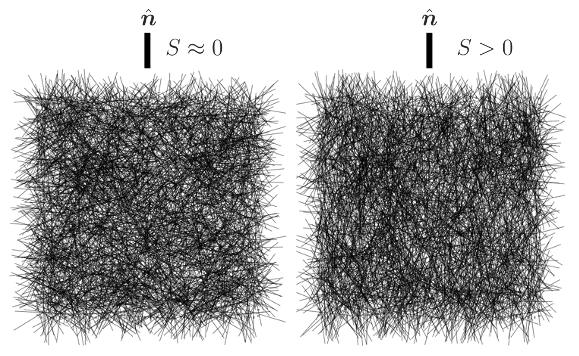
- $S=\sqrt{2 q_{ij} q_{ij}}$ is the nematic order parameter, a scalar quantifying the degree of alignment along $\textbf{n}$ (0: no alignment, i.e. perfect isotropy; 1: complete alignment).
Rather than quantifying $\mathbf{n}$ and $S$ throughout the steps of a simulation, here we want to leverage Cytosim's capabilities in order to attain prescribed nematic characteristics for a filament ensemble.
Theory¶
As discussed in greater detail in an upcoming paper, we introduce a system-wide restraining energy of stiffness $\mathcal{K}_S$ that penalizes deviations of the filament nematic ordering from the one that we intend to prescribe.
For a discrete system comprised of $N$ segments whose direction is described by the unit vectors $m_i^I$, the current nematic tensor reads:
$$ q_{ij}^{curr} = \frac{1}{N} \sum_{I=1}^{N}{m_i^I m_j^I - \delta_{ij}/2} $$where $m_i^I$ denotes the unit vector corresponding to the $I-$th segment that is part of the considered ensemble.
On the other hand, the target nematic tensor is:
$$ q_{ij}^{tgt} = \frac{S_0}{2} \left( n_i n_j - r_i r_j \right) $$where $r_i$ denotes a unit vector that lies in the plane of the system and is directed along the normal to $n_i$.
The restraining energy is thus expressed as
$$ E_S = \frac{\mathcal{K}_S}{2} \left( q_{ij}^{curr} - q_{ij}^{tgt} \right) \left( q_{ij}^{curr} - q_{ij}^{tgt} \right) $$and will result in restraining forces that act on the filament nodes in order to enforce the prescribed nematic ordering. For a generic node of position $X_i^P$, the restraining force reads:
$$ f_i^P = - \frac{\partial E_S}{\partial X_i^P} $$Carrying out the mathematical derivations and considering the node of position $X_i^P$ as identified by the connection of two segments with directions $$m_i^{P,-} = \frac{X_i^{P} - X_i^{P-1} } {\sqrt{(X_i^{P} - X_i^{P-1})(X_i^{P} - X_i^{P-1})}}$$ $$m_i^{P,+} = \frac{X_i^{P+1} - X_i^{P} } {\sqrt{(X_i^{P+1} - X_i^{P})(X_i^{P+1} - X_i^{P})}}$$
we reach the following expression
$$ f_i^P = - \; \frac{\mathcal{K}_S}{N} \left( {q}^{curr}_{ij} - {q}^{curr}_{ij} \right) \left(\begin{array}{c} \frac{\partial m_i^{P,-}}{\partial X_i^P} m_j^{P,-} + m_i^{P,-} \frac{\partial m_j^{P,-}}{\partial X_i^P} + \frac{\partial m_i^{P,+}}{\partial X_i^P} m_j^{P,+} + m_i^{P,+} \frac{\partial m_j^{P,+}}{\partial X_i^P} \\ \frac{\partial m_i^{P,-}}{\partial X_j^P} m_j^{P,-} + m_i^{P,-} \frac{\partial m_j^{P,-}}{\partial X_j^P} + \frac{\partial m_i^{P,+}}{\partial X_j^P} m_j^{P,+} + m_i^{P,+} \frac{\partial m_j^{P,+}}{\partial X_j^P} \end{array}\right)$$which can be further simplified for points that coincide with the fiber ends, since either $m_i^{P,+}$ or $m_i^{P,-}$ will not exist in this case (derivation not shown).
For filament that are represented by segments of constant length $s$ (i.e. the segmentation parameters in Cytosim), the derivatives of the unit vectors appearing above are obtained directly from the definitions of $m_i^{P,-}$ and $m_i^{P,+}$:
$$ \begin{aligned} \frac{ \partial m_i^{P,-}}{\partial X_i^P} = \frac{\left( X^P_j - X^{P-1}_j \right)^2}{s^3}; \;\;\;\;\;\; & \frac{ \partial m_j^{P,-}}{\partial X_i^P} = - \frac{\left( X^P_i - X^{P-1}_i \right)\left( X^P_j - X^{P-1}_j \right)}{s^3} \\ \frac{ \partial m_i^{P,-}}{\partial X_j^P} = - \frac{\left( X^P_i - X^{P-1}_i \right)\left( X^P_j - X^{P-1}_j \right)}{s^3}; \;\;\;\;\;\; & \frac{ \partial m_j^{P,-}}{\partial x_j^P} = \frac{\left( X^P_i - X^{P-1}_i \right)^2}{s^3} \\ \frac{ \partial m_i^{P,+}}{\partial X_i^P} = - \frac{\left( X^{P+1}_j - X^{P}_j \right)^2}{s^3}; \;\;\;\;\;\; & \frac{ \partial m_j^{P,+}}{\partial X_i^P} = \frac{\left( X^{P+1}_i - X^{P}_i \right)\left( X^{P+1}_j - X^{P}_j \right)}{s^3} \\ \frac{ \partial m_i^{P,+}}{\partial X_j^P} = \frac{\left( X^{P+1}_i - X^{P}_i \right)\left( X^{P+1}_j - X^{P}_j \right)}{s^3}; \;\;\;\;\;\; & \frac{ \partial m_j^{P,+}}{\partial X_j^P} = - \frac{\left( X^{P+1}_i - X^{P}_i \right)^2}{s^3} \\ \end{aligned}$$Cytosim implementation and example¶
We implement the described restraining energy as a dedicated Cytosim organizer, which collects an ensemble of filaments with prescribed properties and can be accessed as shown below:
cytosim
set nematic actin_set
{
length = 1.3
stiffness = 5000
direction = 0,1
nematicS = 0.3
}
new 1 actin_set
{
fibers = 3000, filament
placement = none
}This class takes the following parameters:
- length $\rightarrow$ the length of each fiber comprised in the modeled ensemble;
- stiffness $\rightarrow$ the stiffness of the restraining energy, $\mathcal{K}_s$;
- direction $\rightarrow$ the components of the nematic director $\mathbf{n}$;
- nematicS $\rightarrow$ the target value of the nematic order parameter, $S_0$, defined between $0$ and $1$.
As evident, we will need to define separately the properties of the filaments that we want to use in the organizer, whose name and number are passed using the keyword "fibers".
To showcase the functionality of the implemented organizer, we consider a system of actin filaments and modify its nematic ordering throughout the simulation (see image). An corresponding example input file is provided in "nematic_control.cym".